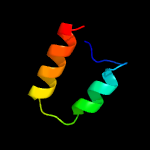
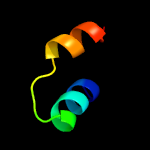
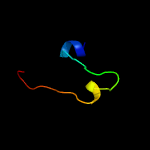
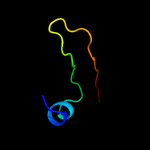
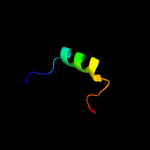
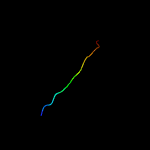
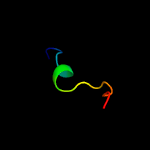
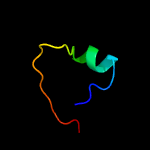
1 d1q8ra_
99.6
32
Fold: Bacillus chorismate mutase-likeSuperfamily: Holliday junction resolvase RusAFamily: Holliday junction resolvase RusA
2 c6idyC_
33.3
26
PDB header: lipid binding proteinChain: C: PDB Molecule: lipase aflb;PDBTitle: crystal structure of aspergillus fumigatus lipase b
3 c5zc1A_
26.4
21
PDB header: hydrolase inhibitorChain: A: PDB Molecule: cystatin-1;PDBTitle: x-ray diffraction analysis of the csstefin-1
4 c7crcB_
23.0
15
PDB header: plant proteinChain: B: PDB Molecule: nad+ hydrolase (nadase);PDBTitle: cryo-em structure of plant nlr rpp1 tetramer in complex with atr1
5 c3vh5D_
20.0
63
PDB header: dna binding proteinChain: D: PDB Molecule: cenp-x;PDBTitle: crystal structure of the chicken cenp-t histone fold/cenp-w/cenp-2 s/cenp-x heterotetrameric complex, crystal form i
6 c3w06A_
12.7
19
PDB header: hydrolaseChain: A: PDB Molecule: hydrolase, alpha/beta fold family protein;PDBTitle: crystal structure of arabidopsis thaliana dwarf14 like (atd14l)
7 c4wnxA_
12.4
24
PDB header: laminin binding proteinChain: A: PDB Molecule: netrin-4;PDBTitle: netrin 4 lacking the c-terminal domain
8 d1hywa_
12.0
30
Fold: gpW/XkdW-likeSuperfamily: Head-to-tail joining protein W, gpWFamily: Head-to-tail joining protein W, gpW
9 c6u08H_
11.5
37
PDB header: toxinChain: H: PDB Molecule: dddi;PDBTitle: double-stranded dna-specific cytidine deaminase type vi secretion2 system effector and cognate immunity complex from burkholderia3 cenocepacia
10 c4ploA_
11.3
48
PDB header: protein bindingChain: A: PDB Molecule: netrin-1;PDBTitle: crystal structure of chicken netrin-1 (ln-le3) in complex with mouse2 dcc (fn4-5)
11 c4aqsA_
11.1
24
PDB header: cell adhesionChain: A: PDB Molecule: laminin subunit beta-1;PDBTitle: laminin beta1 ln-le1-4 structure
12 c4khbF_
10.3
38
PDB header: transcription/replicationChain: F: PDB Molecule: uncharacterized protein pob3n;PDBTitle: structure of the spt16d pob3n heterodimer
13 d1e5qa2
10.0
30
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Homoserine dehydrogenase-like
14 c6losH_
8.9
35
PDB header: signaling proteinChain: H: PDB Molecule: collagen model peptide, type i, alpha 1;PDBTitle: crystal structure of mouse pedf in complex with heterotrimeric2 collagen model peptide.
15 c3b6nA_
8.8
24
PDB header: lyaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase;PDBTitle: crystal structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate2 synthase pv003920 from plasmodium vivax
16 c5gn0H_
8.5
42
PDB header: transcriptionChain: H: PDB Molecule: ww domain-containing transcription regulator protein 1;PDBTitle: structure of taz-tead complex
17 c6m9kD_
8.4
25
PDB header: hydrolaseChain: D: PDB Molecule: recombination protein bet;PDBTitle: crystal structure of lambda exonuclease in complex with the red beta2 c-terminal domain
18 c6losG_
7.7
38
PDB header: signaling proteinChain: G: PDB Molecule: collagen model peptide, type i, alpha 1;PDBTitle: crystal structure of mouse pedf in complex with heterotrimeric2 collagen model peptide.
19 c2ezeA_
7.5
40
PDB header: dna binding protein/dnaChain: A: PDB Molecule: high mobility group protein hmg-i/hmg-y;PDBTitle: solution structure of a complex of the second dna binding2 domain of human hmg-i(y) bound to dna dodecamer containing3 the prdii site of the interferon-beta promoter, nmr, 354 structures
20 d1dgsa2
7.4
23
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA ligase/mRNA capping enzyme postcatalytic domain
21 c4khbE_
not modelled
6.6
26
PDB header: transcription/replicationChain: E: PDB Molecule: uncharacterized protein spt16d;PDBTitle: structure of the spt16d pob3n heterodimer
22 c2xuaH_
not modelled
6.3
21
PDB header: hydrolaseChain: H: PDB Molecule: 3-oxoadipate enol-lactonase;PDBTitle: crystal structure of the enol-lactonase from burkholderia2 xenovorans lb400
23 c2k4fA_
not modelled
6.2
32
PDB header: immune system, signaling proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd3 epsilon chain;PDBTitle: mouse cd3epsilon cytoplasmic tail
24 c7c9r9_
not modelled
6.0
45
PDB header: photosynthesisChain: 9: PDB Molecule: alpha subunit 2 of light-harvesting 1 complex;PDBTitle: structure of photosynthetic lh1-rc super-complex of thiorhodovibrio2 strain 970
25 c4r33A_
not modelled
6.0
26
PDB header: lyaseChain: A: PDB Molecule: nosl;PDBTitle: x-ray structure of the tryptophan lyase nosl with tryptophan and s-2 adenosyl-l-homocysteine bound
26 d1fx7a3
not modelled
6.0
40
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: FeoA-like
27 d1cf7b_
not modelled
5.8
36
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Cell cycle transcription factor e2f-dp
28 c6k31B_
not modelled
5.6
43
PDB header: lyaseChain: B: PDB Molecule: aipepck;PDBTitle: crystal structure of pyrophosphate-dependent phosphoenolpyruvate2 carboxykinase (ppi-pepck)
29 c4aqtA_
not modelled
5.4
29
PDB header: cell adhesionChain: A: PDB Molecule: laminin subunit gamma-1;PDBTitle: laminin gamma1 ln-le1-2 structure
30 c6x61H_
not modelled
5.3
40
PDB header: transcription/transferaseChain: H: PDB Molecule: adaptive-response sensory-kinase sasa;PDBTitle: crystal structure of the n-terminal thioredoxin domain of sasa in2 complex with the n-terminal ci domain of kaic from thermosynchococcus3 elongatus
31 d1zo0a1
not modelled
5.2
44
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: Ornithine decarboxylase antizyme-like
32 c2y38A_
not modelled
5.1
29
PDB header: structural proteinChain: A: PDB Molecule: laminin subunit alpha-5;PDBTitle: laminin alpha5 chain n-terminal fragment
33 d1pkpa1
not modelled
5.1
38
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components
34 c2rsyB_
not modelled
5.1
28
PDB header: transferase/signaling proteinChain: B: PDB Molecule: phosphoprotein associated with glycosphingolipid-enrichedPDBTitle: solution structure of the sh2 domain of csk in complex with a2 phosphopeptide from cbp